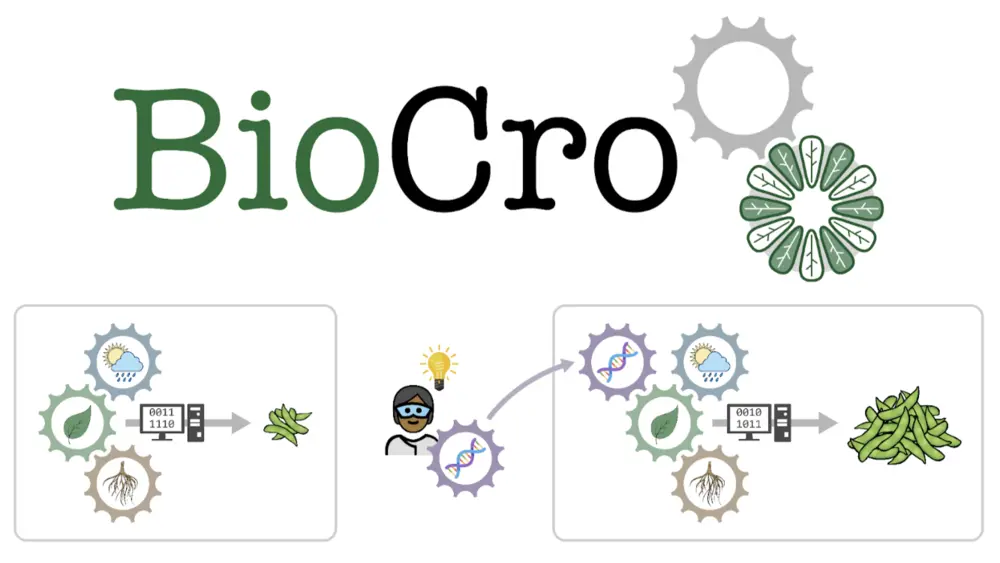
“In the original BioCro, all the math that the modelers were using was mixed into the programming language, which many people weren’t familiar with, so it was easy to make mistakes,” said Justin McGrath, a Research Plant Physiologist for the U.S. Department of Agriculture, Agricultural Research Service (USDA-ARS) at Illinois. “BioCro II separates those so modelers can do less programming and can instead focus on the equations.”
Separating the equations from the programming language allows researchers to try new simulations more easily. For example, if a project is looking at how a gene can help plants to use light more efficiently, the equations for that specific gene can be added to existing models, rather than having to change the entire model to include the new information. This development also allows for the software to operate well with other models, a large improvement from the original BioCro.
In a recent study, published in in silico Plants, McGrath and his team discuss all the improvements they made to the original BioCro software, and why they were necessary to improve modeling capabilities for researchers.
“If you’ve got a gene and you’re wondering how much it can improve yield, you have a tiny piece in the context of the whole plant. Modeling lets you take that one change, put it in the plant and compare yield with and without that change,” said Edward Lochocki, lead author on the paper and postdoctoral researcher for RIPE. “With the updates we’ve made in BioCro II, if you have ten gene changes to make, you can look at all of them quickly and gauge relative importance before moving the work into the field.”